Protocols for E0577
Rapid PNGase F Release of Antibody Glycans
For optimal heat transfer, use 0.2 ml thin wall microtubes or alternatively, 1.5 ml centrifuge tubes. A thermal cycler with heated lid, or a microtube heat block, are suitable for incubation.
One-step Protocol:
- Combine up to 100 μg of antibody and H2O to a volume of 16 μl.
- Add 4 μl of Rapid PNGase F Buffer (5X) to make a 20 μl total reaction volume.
- Add 1 μl of Rapid PNGase F.
- Incubate 10 minutes at 50°C.
Two-step Protocol:
Some antibodies (i.e., Fab N-glycans) require a preheating step for efficient deglycosylation.
- Combine up to 100 μg of antibody and H2O to a volume of 16 μl.
- Add 4 μl of Rapid PNGase F Buffer (5X) to make a 20 μl total reaction volume.
- Incubate at 80°C for 2 minutes, cool down.
- Add 1 μl of Rapid PNGase F.
- Incubate 10 minutes at 50°C.
Fluorescent Labeling with Procainamide (PCA), or 2-aminobenzamide (2AB)
- Add 18 µl of acidified PCA or 2AB labeling reagent and 24 µl cyanoborohydride reagent to the deglycosylation reaction and incubate for 45 minutes at 65°C.
Note: Stock solutions can be made fresh or kept at -20°C and thawed prior to use (reagents remain stable for several weeks and through numerous freeze/thaw cycles). Reagents prepared as follows: PCA (550 mg dissolved in 1 ml DMSO), 2AB (250 mg dissolved in 1 ml DMSO), and sodium cyanoborohydride (200 mg/ml in H2O). Prepare acidified PCA or 2AB solution by adding one volume of glacial acetic acid to eight volumes of PCA or 2AB stock solution.
- Cool reactions to room temperature.
Glycan Purification with a HILIC Spin Column
Released, labeled glycans can be purified from the free label using various methods. Purified glycans must be free of organic solvents prior to enzyme digestion.
- Add 350 µl Acetonitrile (ACN) to the labelled reactions to a final concentration of 85% ACN.
- Using either a vacuum manifold or centrifuge adapter (following manufacturer’s instructions), condition a HILIC spin column with 350 µl H2O, followed by 350 µl of 50 mM ammonium formate, pH 4.5.
- Equilibrate column with 350 µl of 85% ACN/15% ammonium formate.
- Load sample onto the HILIC column, spin.
- Wash column with 300 µl of 1% formic acid, 90% ACN, repeat 5 times.
- Elute glycans into a fresh collection tube with 30 µl of 50 mM ammonium formate, pH 4.4. Repeat 3 times for a final volume of 90 µl.
- Dry the 90 µl glycan sample in a speed vac with temperature no greater than 4°C or lyophilize overnight Note: To minimize drying time, samples can be aliquoted into several tubes prior to placing in speed vac.
- Resuspend the sample in 100 µl H2O for subsequent exoglycosidase digestion reactions. The resuspension volume can be modified to accommodate subsequent exoglycosidase reactions.
Exoglycosidase Digestion of Labelled Glycans
Exoglycosidase reactions can be assembled with single or multiple enzymes. The amount of labeled glycan should be determined based on the downstream method of analysis (i.e., HPLC, UPLC, MS, CE). Considerations include the instrument range of detection and the signal to noise ratio of the fluorophore used. This protocol allows for the digestion of up to 130 pmol (equivalent to 10 µg of antibody) using the recommended amounts of exoglycosidase(s).
- Combine up to 130 pmol of labeled, purified glycans (10 µg antibody) and H2O to a total reaction volume of 20 µl.
Note: Labeled glycans can be increased to a maximum of 330 pmol (25 µg of antibody) using the recommended amount of exoglycosidases with an incubation time of 18 hr at 37°C.
- Add 2 µl of 10X GlycoBuffer 1.
- Add the recommended volume of each exoglycosidase (single or in combination) to yield the specific structures, as shown in Table 1.
- Incubate reactions for 3 hours at 37°C. Note: If digestion is not complete, increase incubation time to 18 hours at 37°C.
Samples can be analyzed directly with MS or CE or can be further purified to remove enzymes from the reactions if necessary.
Table 1: Exoglycosidase Digestion Panel

Example 1: Characterization of Infliximab Glycans using Exoglycosidase Panel
Infliximab (30 µg) was deglycosylated with Rapid PNGase F, released glycans were labeled with procainamide (PCA) and purified as described in the general protocol. PCA-labeled glycans were lyophilized and resuspended in 30 µl of water.
- Prepare exoglycosidase reactions as described in Table 2:
Table 2: Exoglycosidase Digestion Panel

- Incubate samples for 3 hours at 37°C
- Add 10 μl of 50 mM NH4 formate buffer pH 4.4 and 90 μl acetonitrile to each reaction for a final acetonitrile concentration of 70%.
- Analyze by LCMS, results shown in Figure 3A and 3B: N-glycan samples are separated using a XBridge® BEH Amide column (Waters) on a Dionex® UltiMate® LC equipped with fluorescent detection in line with a LTQ® Orbitrap® Velos Spectrometer equipped with a heated electrospray standard source (HESI-II probe).

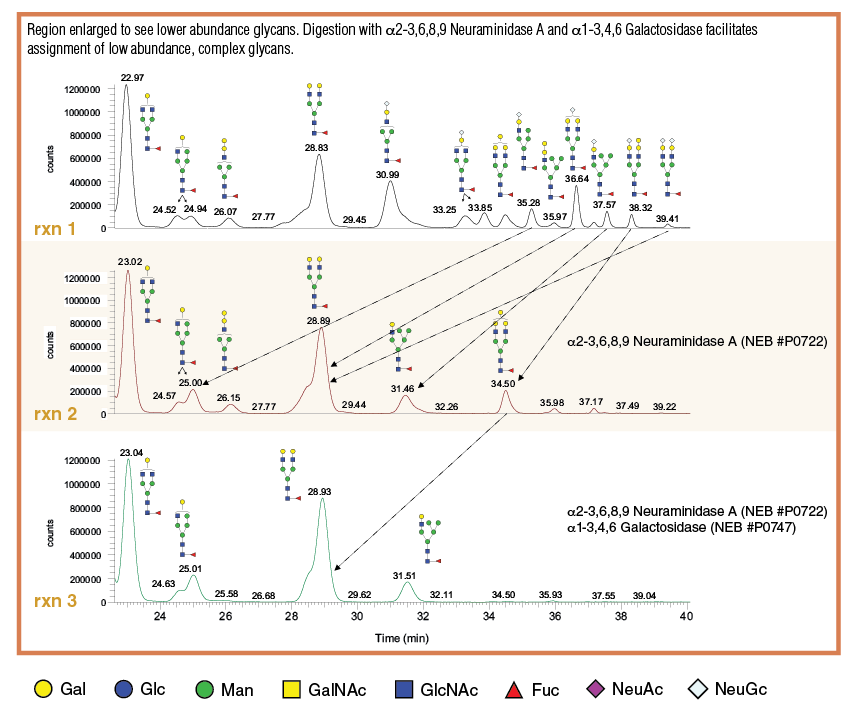
Example 2: Exoglycosidase Combinations to Isolate and Quantitate Potentially Immunogenic Low Abundance Isotopes such as Neu5Gc and a1-3 Galactose in Infliximab, a Murine-derived Therapeutic
Using combinations of enzymes can be useful to simplify the glycan profile data and isolate a species of interest. Glycans were released from 15 µg of Infliximab using Rapid PNGase F and labelled with procainamide (PCA). Purified, labelled glycans were resuspended in 15 µl of H2O.
- Prepare exoglycosidase reactions as described in Table 3:
Table 3: Exoglycosidase Digestion Panel

- Incubate samples for 3 hours at 37°C
- Add 10 μl of 50 mM NH4 formate buffer pH 4.4 and 90 µl acetonitrile to each reaction for a final acetonitrile concentration of 70%.
- Analyze by LCMS, results shown in Figure 4: N-glycan samples are separated using a XBridge BEH Amide column (Waters) on a Dionex UltiMate LC equipped with fluorescent detection in line with a LTQ Orbitrap Velos Spectrometer equipped with a heated electrospray standard source (HESI-II probe).

RXN B: Infliximab glycan digestion with 1 μl of α 1-3,4,6 Galactosidase, 1 μl of β 1-4 Galactosidase S, and 1 μl of β -N-Acetylglucosaminidase S.
RXN C: Infliximab glycan digestion with 2 μl of α 2-3,6,8,9 Neuraminidase A, 1 μl of β 1-4 Galactosidase S, and 1 μl of β -N-Acetylglucosaminidase S.
Example 3: Exoglycosidase Digestion of Enbrel Glycans to Quantitate Overall Level of Fucosylation and High Mannose Structures
Glycans were released from 15 µg of Enbrel using Rapid PNGase F and labelled with procainamide (PCA) as described in the general protocol. Purified, labelled glycans were resuspended in 15 µl of H2O.
- Prepare exoglycosidase reactions as described in Table 4:
Table 4: Exoglycosidase Digestion Panel

- Incubate samples for 3 hours at 37°C
- Add 10 μl of 50 mM NH4 formate buffer pH 4.4 and 90 µl acetonitrile to each reaction for a final acetonitrile concentration of 70%.
- Analyze by LCMS, results shown in Figure 5: N-glycan samples are separated using a XBridge BEH Amide column (Waters) on a Dionex UltiMate LC equipped with fluorescent detection in line with a LTQ Orbitrap Velos Spectrometer equipped with a heated electrospray standard source (HESI-II probe).

RXN B: Enbrel glycan digestion with 2 μl of α 2-3,6,8,9 Neuraminidase λ, 1 μl of β 1-4 Galactosidase S, and 1 μl of β -N-Acetylglucosaminidase S.
RXN C: Enbrel glycan digestion with 2 μl of α 2-3,6,8,9 Neuraminidase λ, 1 μl of β 1-4 Galactosidase S, 1 μl of β -N-Acetylglucosaminidase S, and 2 μl of α 1-2,4,6 Fucosidase O.

