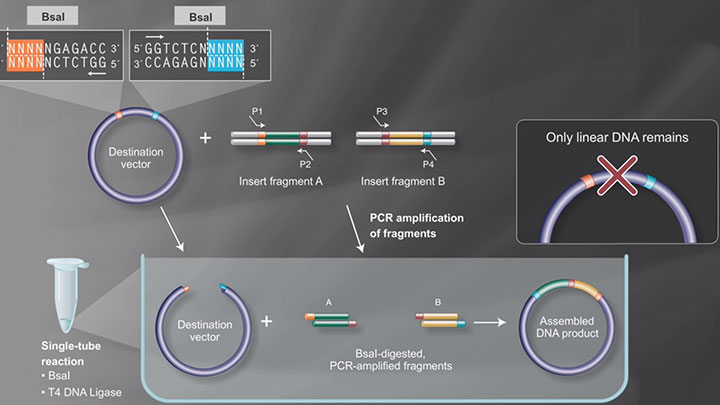
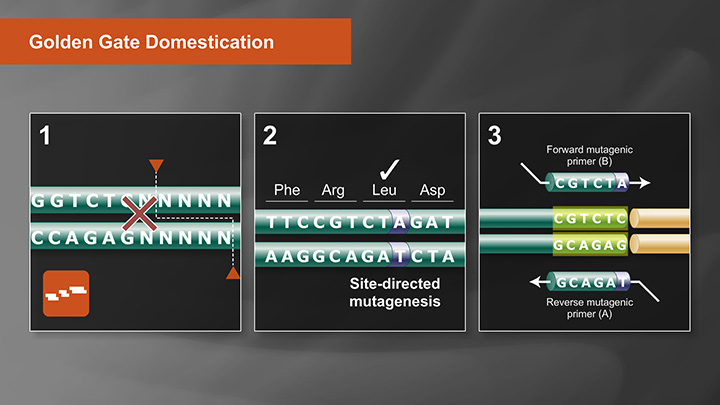
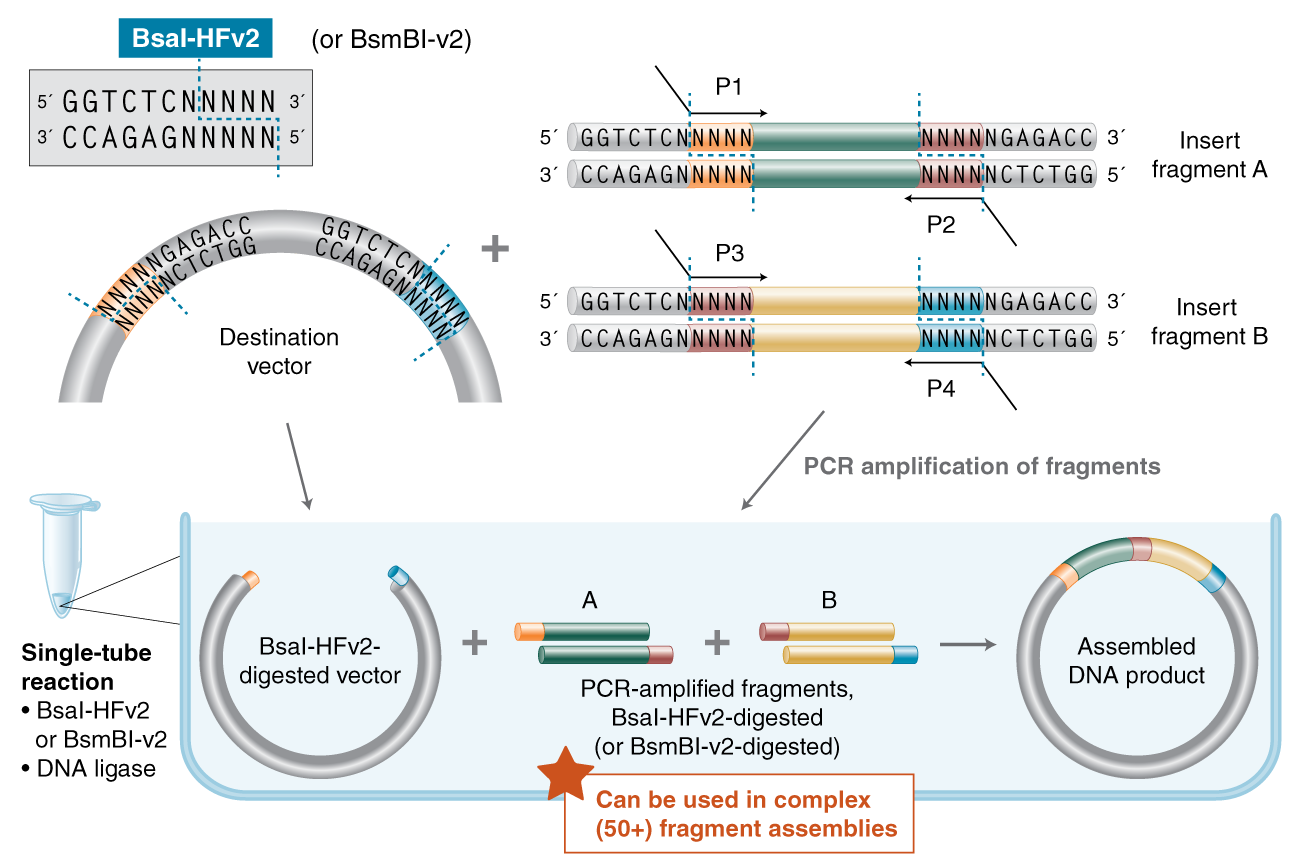
Push the Limits of Golden Gate Assembly
50+ fragment DNA assembly achievable with high efficiency and accuracy!

With constant advances both in the development of new enzymes (e.g., PaqCI®, BsaI-HF®v2 and BsmBI-v2) and research on maximizing enzyme functionality (e.g., ligase fidelity), NEB has become the industry leader in pushing the limits of Golden Gate Assembly (and related methods such as MoClo, GoldenBraid, Mobius Assembly and Loop Assembly). NEB has all the products and information you need to perform complex assemblies.
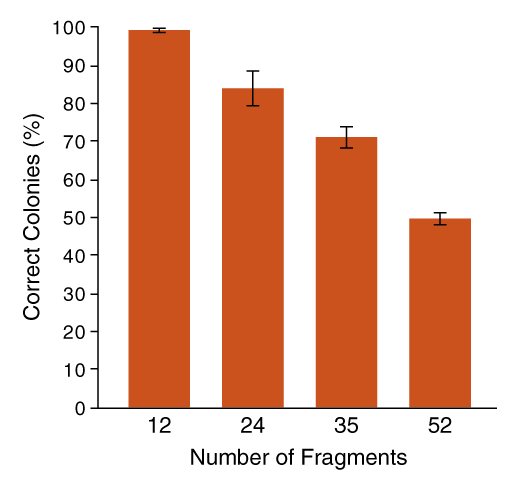
Fidelity decreases as the complexity of Golden Gate Assembly increases. Visit our protocols for multi-fragment assemblies to the right.
Advances in ligase fidelity
Research at NEB has led to an increased understanding of ligase fidelity, including the development of a comprehensive method for profiling end-joining ligation fidelity in order to predict which overhangs have improved fidelity (1, 2, 3, 4). Their research has enabled complex fragment assemblies with high efficiencies and >90% accuracy. Use our Ligase Fidelity Tools to design high fidelity Golden Gate Assemblies under various experimental conditions and learn how these tools and research are enabling advancements in synthetic biology.
Type IIS restriction enzymes for Golden Gate Assembly
NEB offers more Type IIS (i.e., recognize asymmetric DNA sequences and cleave outside of their recognition sequence) restriction enzymes than any other supplier, many of which are used in Golden Gate Assembly. NEB is pleased to offer: Esp3I (an isoschizomer of BsmBI that is recommended for use at 37°C and is supplied with rCutSmart™Buffer), PaqCI (an AarI isoschizomer with a 7 basepair recognition sequence) and the improved BsaI-HFv2 and BsmBI-v2, which are optimized for Golden Gate Assembly.
For information about other Type IIS restriction enzymes used in Golden Gate Assembly, such as PaqCI, BbsI, BbsI-HF and SapI, view our Type IIS restriction enzyme table.
One or more of these products are covered by patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc. For more information, please email us at gbd@neb.com. The use of these products may require you to obtain additional third party intellectual property rights for certain applications.
Featured Products:
| NEBridge® Golden Gate Assembly Kit (BsmBI-v2) Contains an optimized mix of BsmBI-v2 (optimized for Golden Gate Assembly) and T4 DNA Ligase. Can direct the accurate assembly of 2 – 50+ inserts/modules using the Golden Gate approach. |
|
| NEBridge® Ligase Master Mix NEBridge Ligase Master Mix is a 3X master mix for Golden Gate Assembly. Designed for use with NEB Type IIS restriction enzymes, this master mix contains T4 DNA Ligase in an optimized reaction buffer with a proprietary ligation enhancer. |
|
| Esp3I An isoschizomer of BsmBI that is recommended for use at 37°C and is supplied with rCutSmart Buffer, adding flexibility to your reaction setup |
|
| BsaI-HFv2 New version of BsaI-HF that is optimized for Golden Gate Assembly. |
|
| BsmBI-v2 New version of BsmBI that is optimized for Golden Gate Assembly. |
|
| PaqCI New Type IIS restriction enzyme with 7 basepair recognition sequence (AarI isoschizomer). |
Featured Resources
Protocols:
- 13-Fragment Golden Gate Assembly Protocol using SapI (NEB #R0569)
- 24-Fragment Golden Gate Assembly using BsaI-HF®v2 (NEB #R3733)
- 35-Fragment Golden Gate Assembly using BsmBI-v2 (NEB #E1602)
- 52-Fragment Golden Gate Assembly using BsaI-HF®v2 (NEB #E1601)
- Golden Gate Assembly Protocol using PaqCI® (NEB #R0745) and NEBridge Ligase Master Mix (NEB #M1100)
Application Notes:
Golden Gate Assembly Brochure

Read our Golden Gate Assembly brochure for tips, tools and products to help you achieve your complex (50+) fragment assemblies.