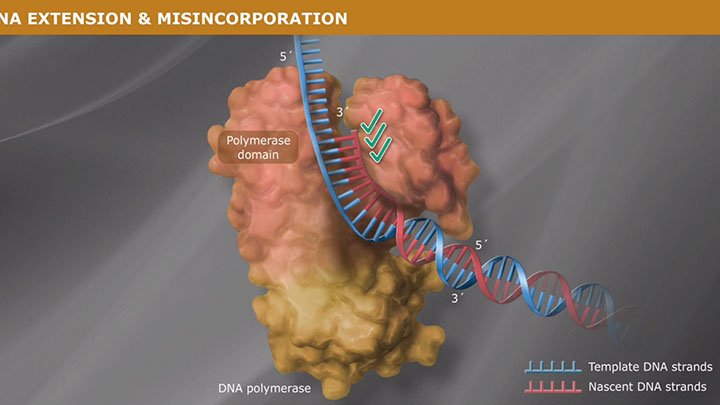
High Fidelity means low error rates, more accurate sequences
Q5 High-Fidelity DNA Polymerase has an ultra-low error rate (<1 error per million bases) due to strong proofreading (3´-5´) exonuclease activity, making it the preferred polymerase for all PCR applications requiring greater accuracy (high sequence accuracy) or the amplification of difficult (from high AT to high GC) templates.
High Processivity means longer amplicons, faster
Q5 High-Fidelity Polymerase is fused to the processivity-enhancing Sso7d DNA binding domain, improving speed, fidelity, and reliability of performance. This increased fidelity and processivity allows for more accurate and longer template (up to 10 kb gDNA, 20 kb plasmid) amplification.
Learn More
Enhanced fidelity
The fidelity of a DNA polymerase is defined by its ability to accurately replicate a template.
A critical aspect of fidelity is the ability of the DNA polymerase to read the template strand, select the appropriate nucleoside triphosphate and insert the correct nucleotide at the 3´ end in the polymerase catalytic domain, such that canonical Watson-Crick base pairing is maintained. High-fidelity polymerases have a significant binding preference for the correct versus the incorrect nucleotide triphosphate during polymerization.
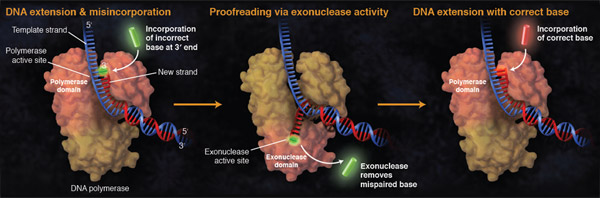
A polymerase's error rate is the rate of misincorporation of an incorrectly matched nucleotide. If an incorrect nucleotide does bind in the polymerase active site, incorporation is slowed due to the sub-optimal architecture of the active site complex. This time provides the opportunity for the incorrect nucleotide to dissociate before incorporation, thereby allowing the process to start again (and for a correct nucleotide triphosphate to bind).
In addition to effective discrimination for correct over incorrect nucleotide incorporation in the polymerase active site, some DNA polymerases possess proofreading (3´→5´) exonuclease activity. If after some time the correct nucleotide is not incorporated, the 3´ end of the growing DNA chain is moved into this 3´→5´ exonuclease domain where the phosphodiester bond between the incorrect nucleotide and the previous nucleotide is broken, releasing the incorrect nucleotide and permitting the chain to move back into the polymerase domain, where polymerization can continue with the correct nucleotide.
Fidelity is important for applications in which the DNA sequence must be correct after amplification, including:
- Cloning/subcloning from in vitro amplified material (PCR, RCA, etc.) for protein expression or gene studies
- SNP analysis by cloning and sequencing
- RNA analysis by RT-PCR
- Next-generation sequencing
Fidelity is less important:
- If the PCR amplified product is directly sequenced by Sanger sequencing (without an intervening cloning step), where the signal is an average of the input amplicons. However, it is more important for next generation and single molecule sequencing techniques.
- In diagnostic applications where the read-out is the presence or absence of a product.
Enhanced processivity by fusion to the Sso7d dsDNA binding protein
Processivity is the ability of a polymerase to replicate DNA without dissociating from the template and is measured by the average number of nucleotides incorporated per binding event. Sso7d binds to the dsDNA, stabilizing the fusion-polymerase to the template, even when the template is difficult.
Sso7d (from Sulfolobus solfataricus) is a 7 kDa, sequence-independent dsDNA binding protein. Sso7d-fusion polymerases allow reduced PCR extension times (limiting nonspecific amplification) and permits the use of less enzyme (cost effective, less storage buffer components, like glycerol) by increasing polymerase processivity.
For example, Pfu (Pyroccocus furiosis) polymerase (an Archaeal Family B polymerase, like Q5®) alone exhibits a processivity score of 0.84 with an average primer extension length of 6 nucleotides, while the fusion protein Pfu-Sso7d exhibits higher processivity with a score of 0.98 and an average extension length of 55 nucleotides (3).
(1) Johnson, K. A. (2010) Biochimica et Biophysica Acta, 1804, 1041–1048. PMID: 20079883
(2) Joyce, C. M., & Benkovic, S. J. (2004) Biochemistry, 43, 14317–14324. PMID: 15533035
(3) Wang, Y. et al. (2004) Nucleic Acids Res, 32, 1197-1207. PMID: 14973201