DNA Library Prep for Illumina
Choose Type:
-
The Quantitation Question: How does accurate library quantitation influence sequencing?
The determination of the number of sequencing-ready molecules present after library preparation is an important step in the next generation sequencing (NGS) workflow and has a strong influence on the success of both a sequencing run and a sequencing-based experiment.
-
Improving Enzymatic DNA Fragmentation for Next Generation Sequencing Library Construction
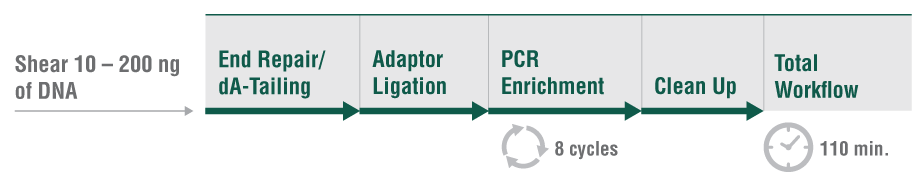
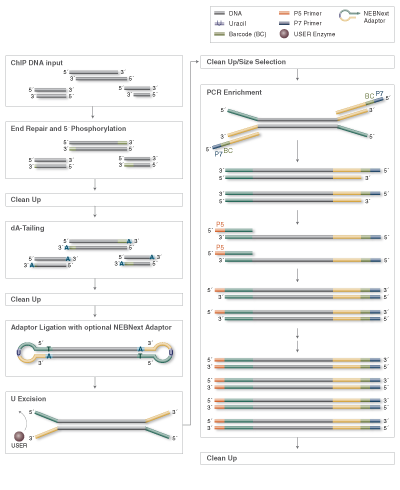
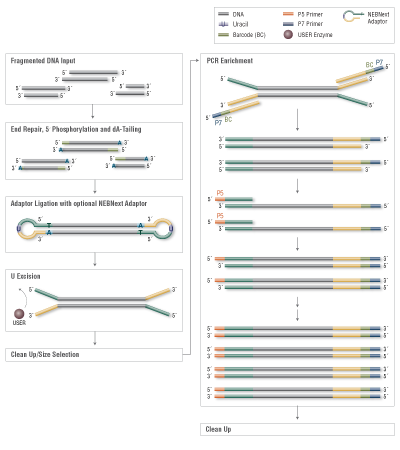
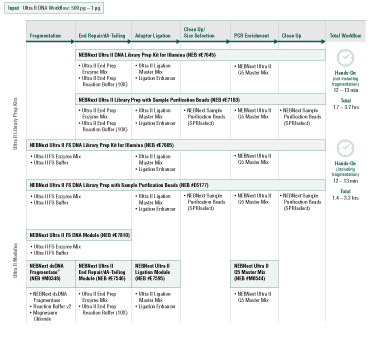
Among the available options for DNA fragmentation, enzymatic fragmentation is demonstrably gentler on the sample, yielding less damage at any scale. The new NEBNext® Ultra™ II FS DNA Library Prep Kit for Illumina® includes a single step for enzymatic fragmentation, end repair, and dA tailing.
- A Robust, Streamlined, Enzyme-based DNA Library Preparation Method Amenable to a Wide Range of DNA Inputs (2019)
- EM-seq™ Enables Accurate and Precise Methylome Analysis of Challenging DNA Samples (2019)
- NEBNext® Ultra™ II FS DNA: A Robust Enzyme-based DNA Library Preparation Method Compatible with Plant Samples
- Uncovering the Cannabis sativa Methylome Through Enzymatic Methyl-seq (2019)
Feature Articles
Posters
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.